- Products
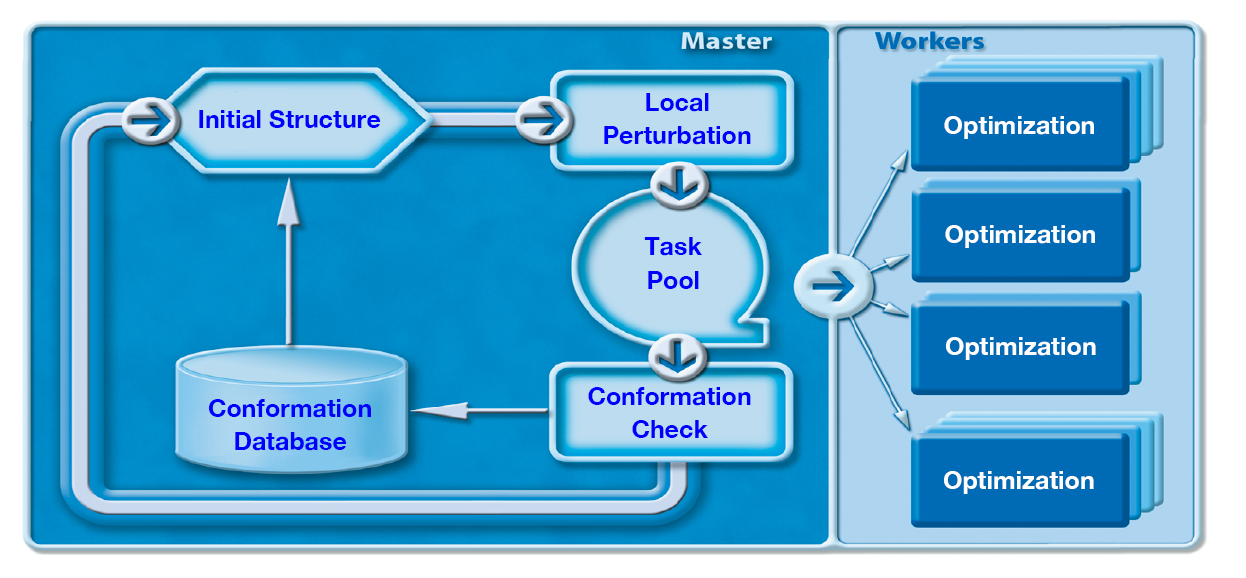
- Parallel CONFLEX
In the conformation search with CONFLEX, the initial structure is deformed by rotations around bonds in chain parts and flap/flip perturbations of atoms that make up a ring in order to generate structures that have various conformations. In the crystal structure search, CONFLEX generates various structures by means of different space groups and initial molecular positions and orientations. While single-core version optimizes the generated structures in turn, parallel version, Parallel CONFLEX, can distribute the optimizations across multiple CPU/cores. Therefore the use of the parallel version can significantly reduce execution time.
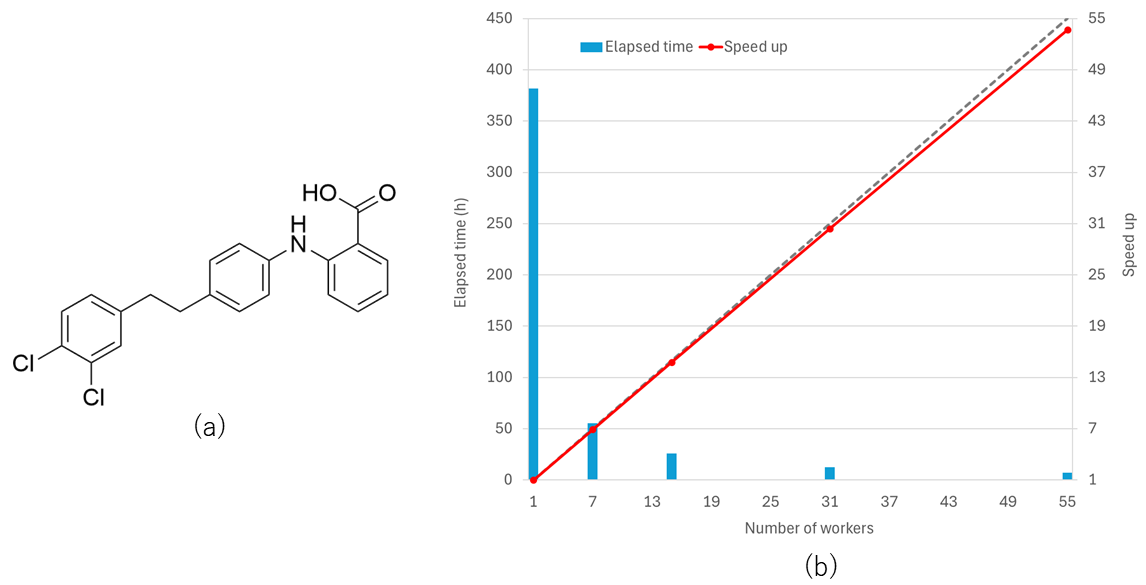
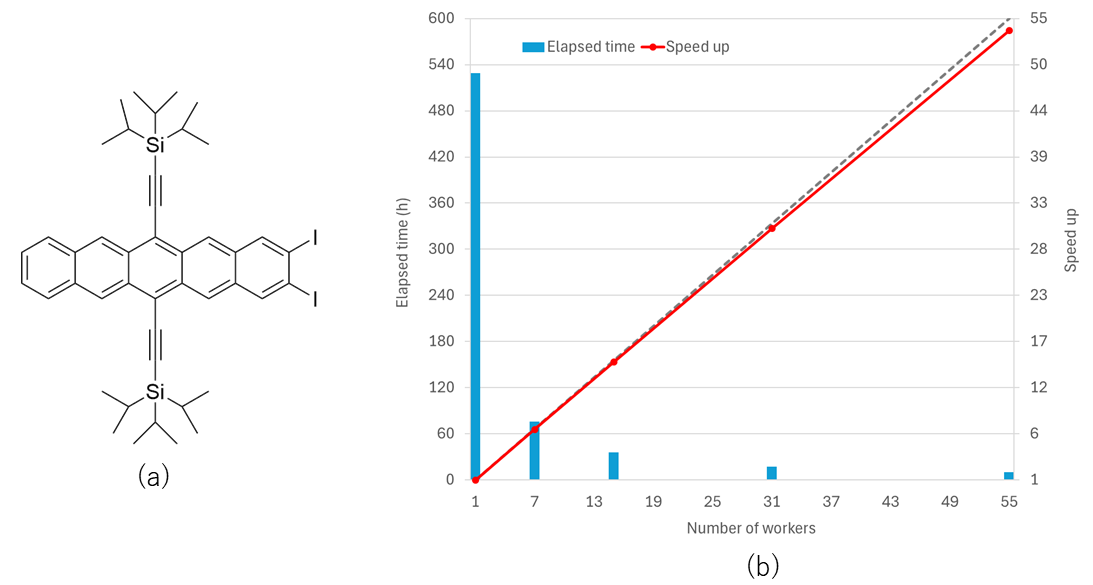
Parallelization performance in the crystal structure search
The figures and table show parallelization performance in the crystal structure search for 3-aza-bicyclo(3.3.1)nonane-2,4-dione A, 2-((4-(3,4-Dichlorophenethyl)phenyl)amino)benzoic acid B, and (2,3-diiodopentacene-6,13-diyl)bis(ethyne-2,1-diyl)]bis(triisopropylsilane) C molecules. As shown in the figures of execution time and speed up for the optimization, we can efficiently reduce execution time by using many workers. The parallelization efficiency is as high as approximately 98 %. In the calculations, Intel(R) Xeon(R) Gold 6258R CPU 2.70 GHz was used.

| Molecule | Number of atoms in asymmetric unit | Number of generated structures | Number of predicted structures | Execution time (h) | Parallelization efficiency (%) |
|---|---|---|---|---|---|
| A | 22 | 104,976 | 1,976 | 4.8 | 99.7 |
| B | 43 | 20,412 | 3,083 | 7.1 | 97.6 |
| C | 100 | 8,748 | 999 | 9.9 | 97.5 |